Multiple Linear Regression with R
Data description:
Analysis:
>ssa=read.table("file=http://ramanujan.math.trinity.edu/ekwess/misc/ssabsorption.txt", header=TRUE) # importing the data
into the R workspace and saving it as "ssa"
>head(ssa) # displaying the first rows
AmountExtraIon AmountExtraAlum PhosphateExtraIndex
>x1=ssa$AmountExtraIon # Renaming the variables
>x2=ssa$AmountExtraAlum
>y=ssa$PhosphateExtraIndex
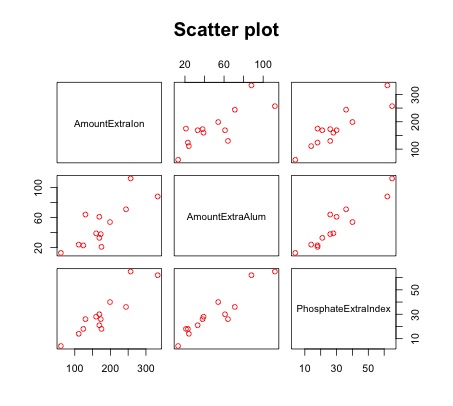
>plot(ssa, main="Scatter Plot",col="red") # Multivariate Scatter plot of y against x1 and x2
>mreg=lm(y~x1+x2) # Performing multiple linear regression of y against the x1 and x2 and saving it as "mreg"
>summary(mreg) # Obtaining the summary of the multiple linera regression
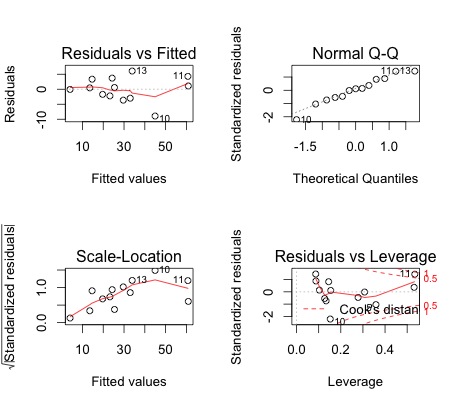
>par(mfrow=c(2,2)) # dividing the plot window into four frames
>plot(mreg) # Residual plots and diagnostics
>par(op) # End dividing.
- The scatter plot shows a linear relationship between the Phosphate Absorption Index, the Amount of Extractable Iron, and the Amount of Extractable Aluminum.
- The summary of the regression suggests a relationship of the form y=-7.35066+0.11273x1+0.34900x2.
- The small p-values (0.003504 and 0.000628) suggest that the coefficient estimates are highly significant.
- The coefficient of determination (0.9382) suggests that about 94% of change in the Phosphate Absorption Index is due to changes in the Amounts of Extractable Iron and Aluminum respectively.
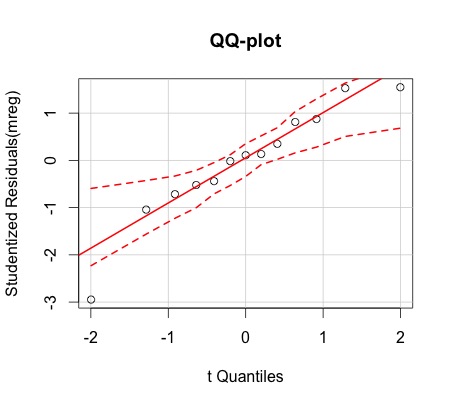
- The normal QQ plots shows that the assumption of normality is valid. The residuals-vs-fitted values-plot does not show a particular pattern, and the vertical spread does not vary too along the horizontal lenght. Moreover, this plot also suggests that the residuals may be centered around 0. Thus it is likely that the assumptions of independence, equal mean 0, and that of equal variances (homoscedasticity) are satisfied.
- There are ways to check some of these assumptions individually in R.
Checking Assumptions:
>install.packages("nortest") # Instaling the package "nortest"\
>require(nortest) # Loading the package nortest into the R workspace
>res<-mreg$resid # saving the residuals from the regression as "res"
>ad.test(res) # Anderson-Darling test of normality
>cvm(res) # Cramer-Von-Mises test of normality
>lillie.test(res) # Kolomogorov-Smirnov test of normality
>sf.test(res) # Shapiro-Francia test of normality
>shapiro.test(res) # Shapiro test of normalty
>pearson.test(res) # Pearson test of normality.
-QQ-plot
>qqp(mreg, envelope=0.95, main="QQ-plot") # QQplot with an envelope showing a 95% point-wise confidence level.
>require(car) # Loading the package "car"
> ncvTest(mreg) # R tests if the errors have non constant variance.
>durbinWatsonTest(mreg) # R tests for autocorrelation between the errors. The required packages is "car"